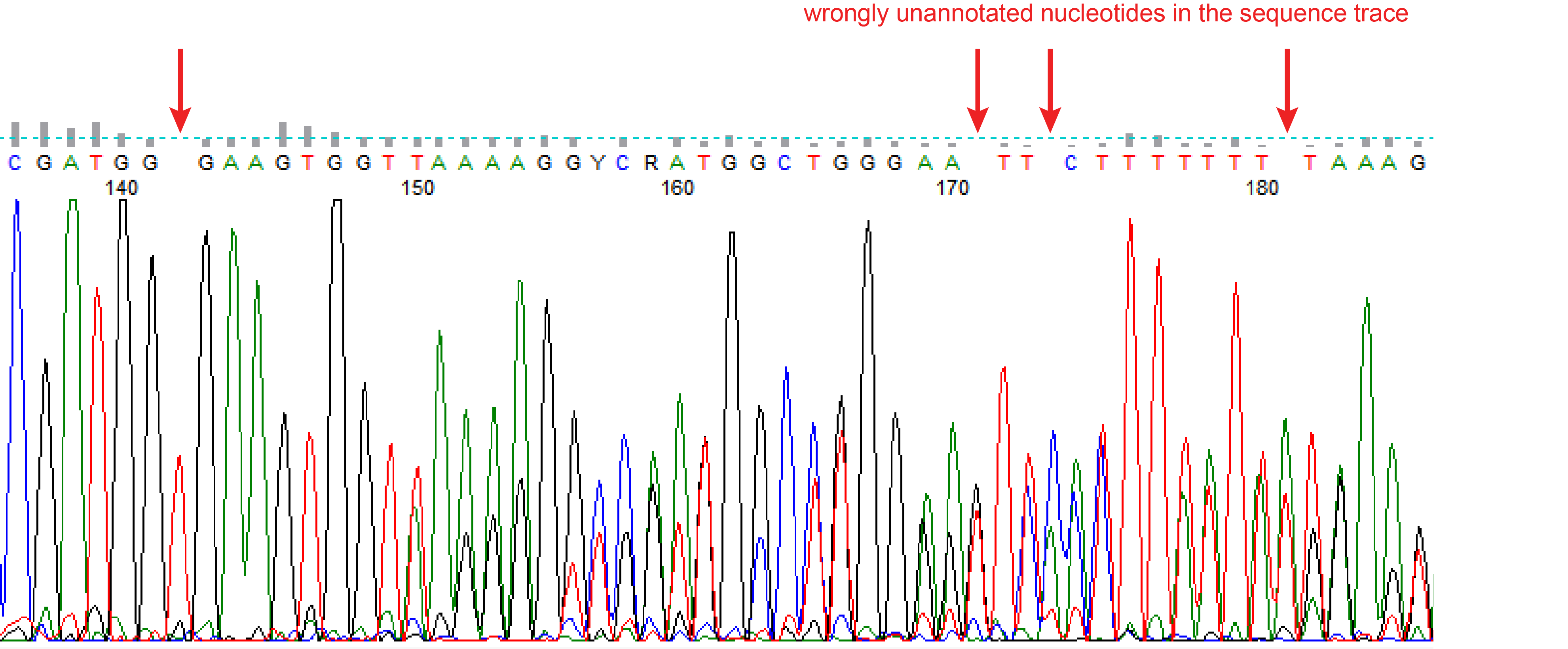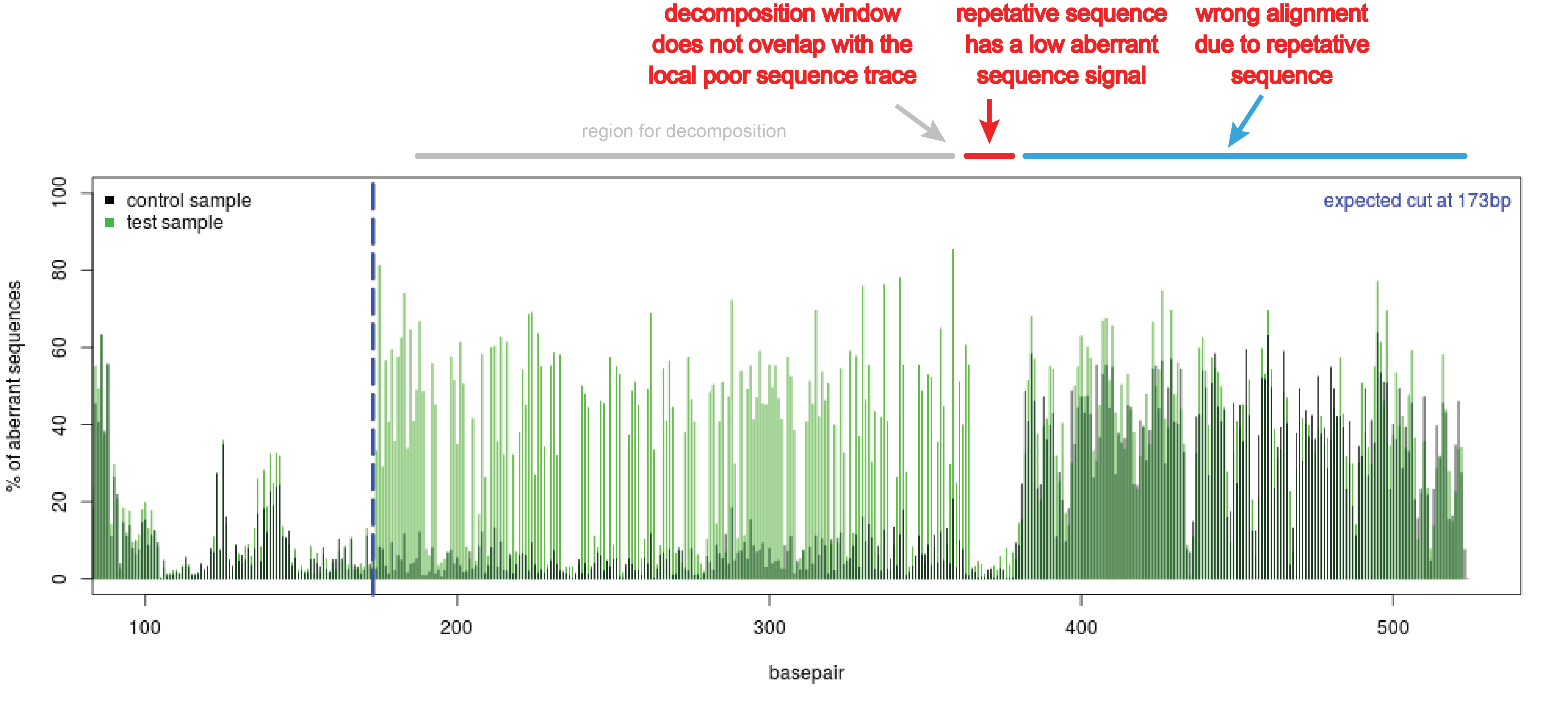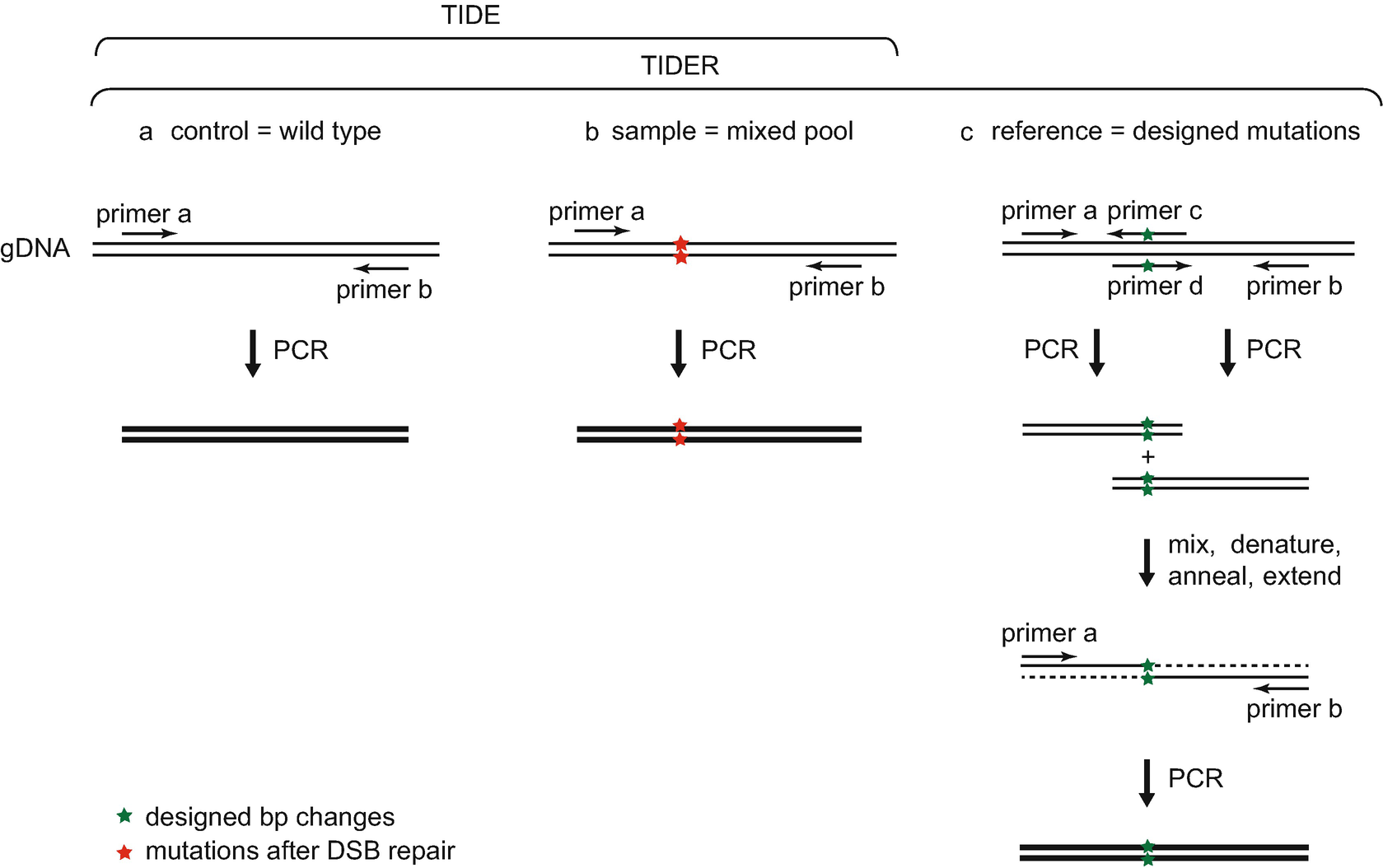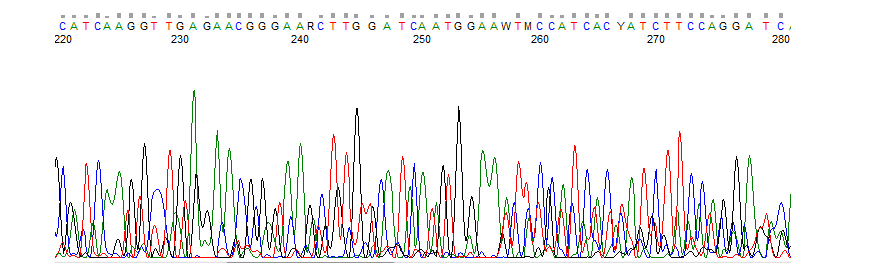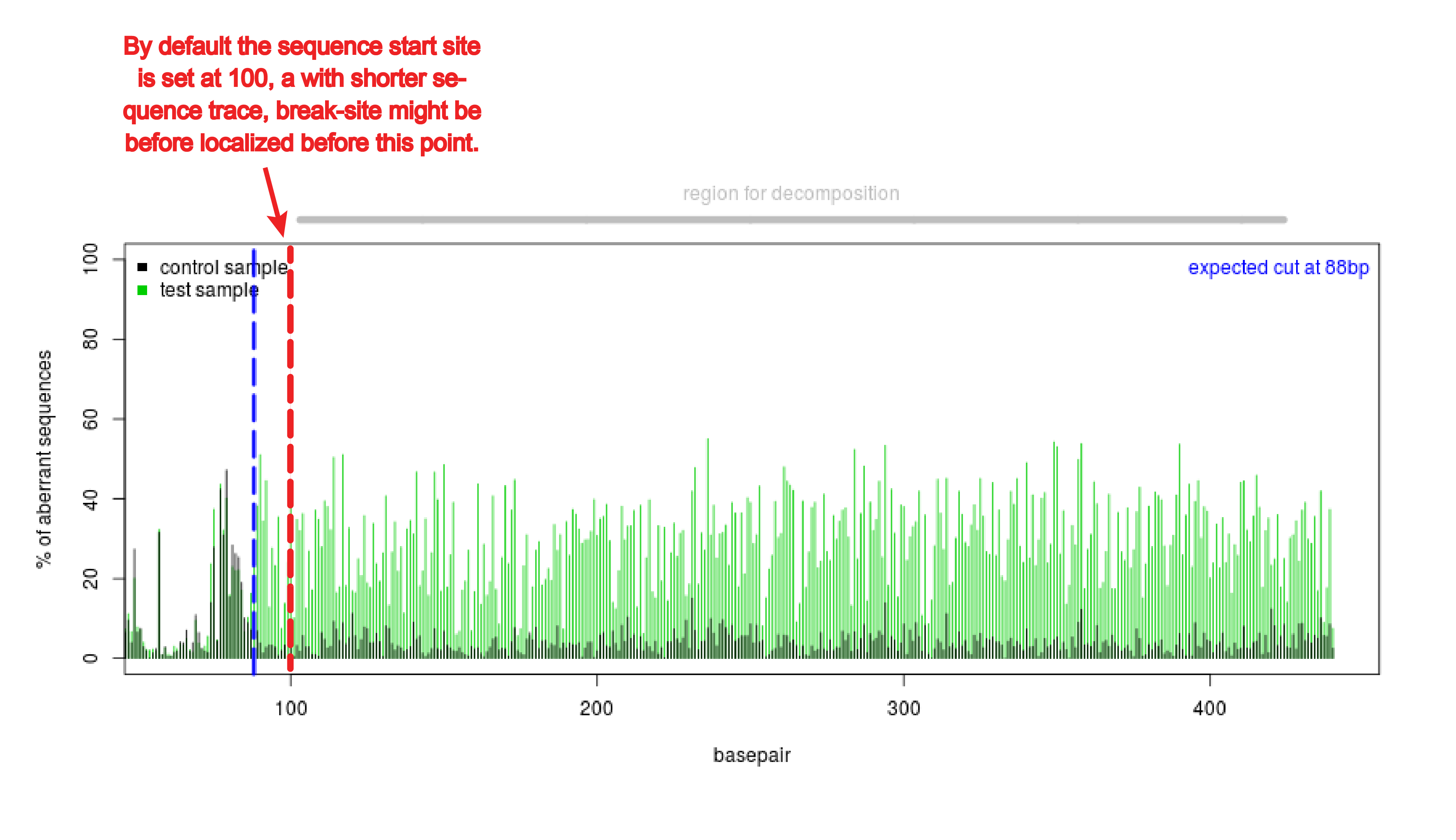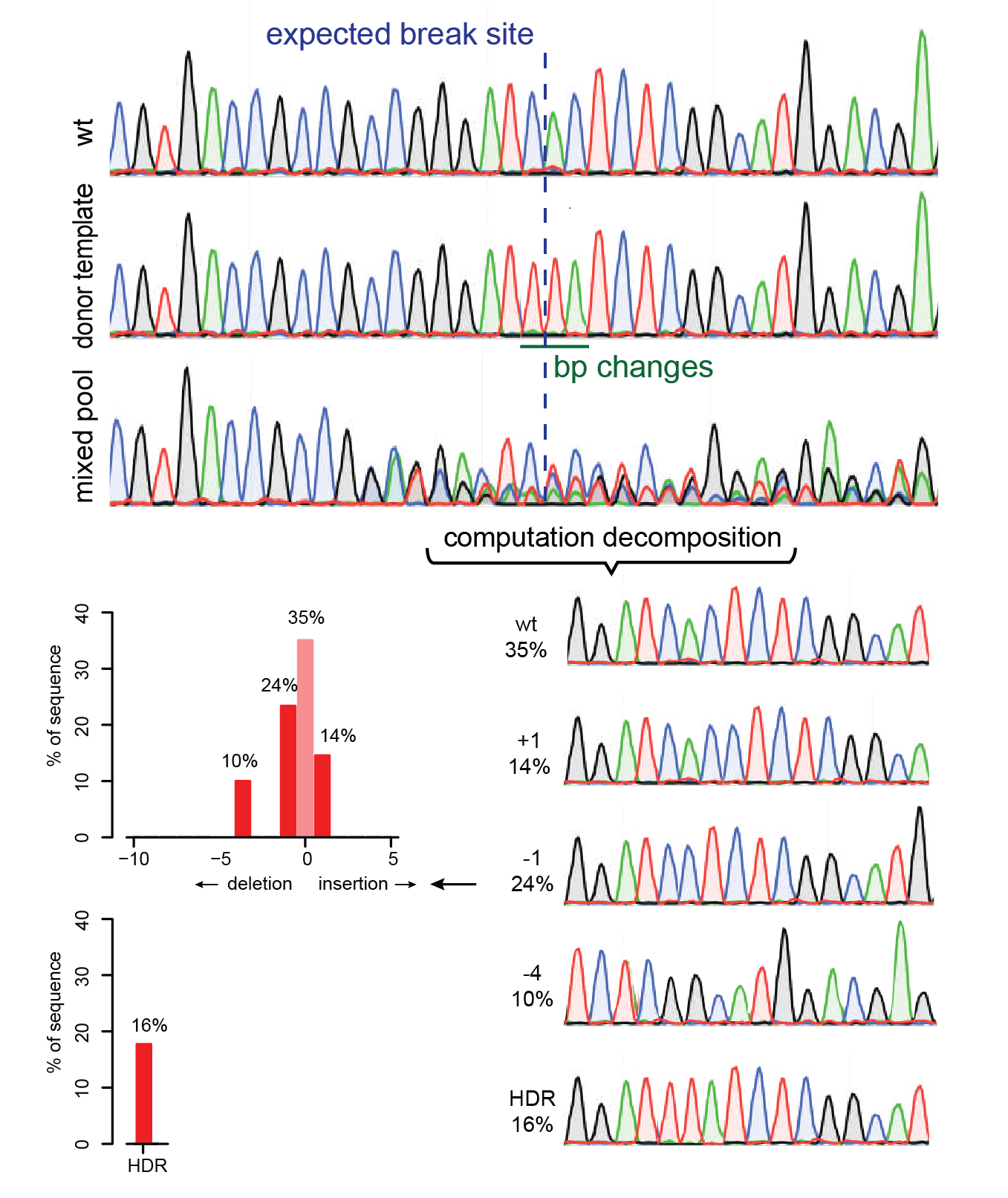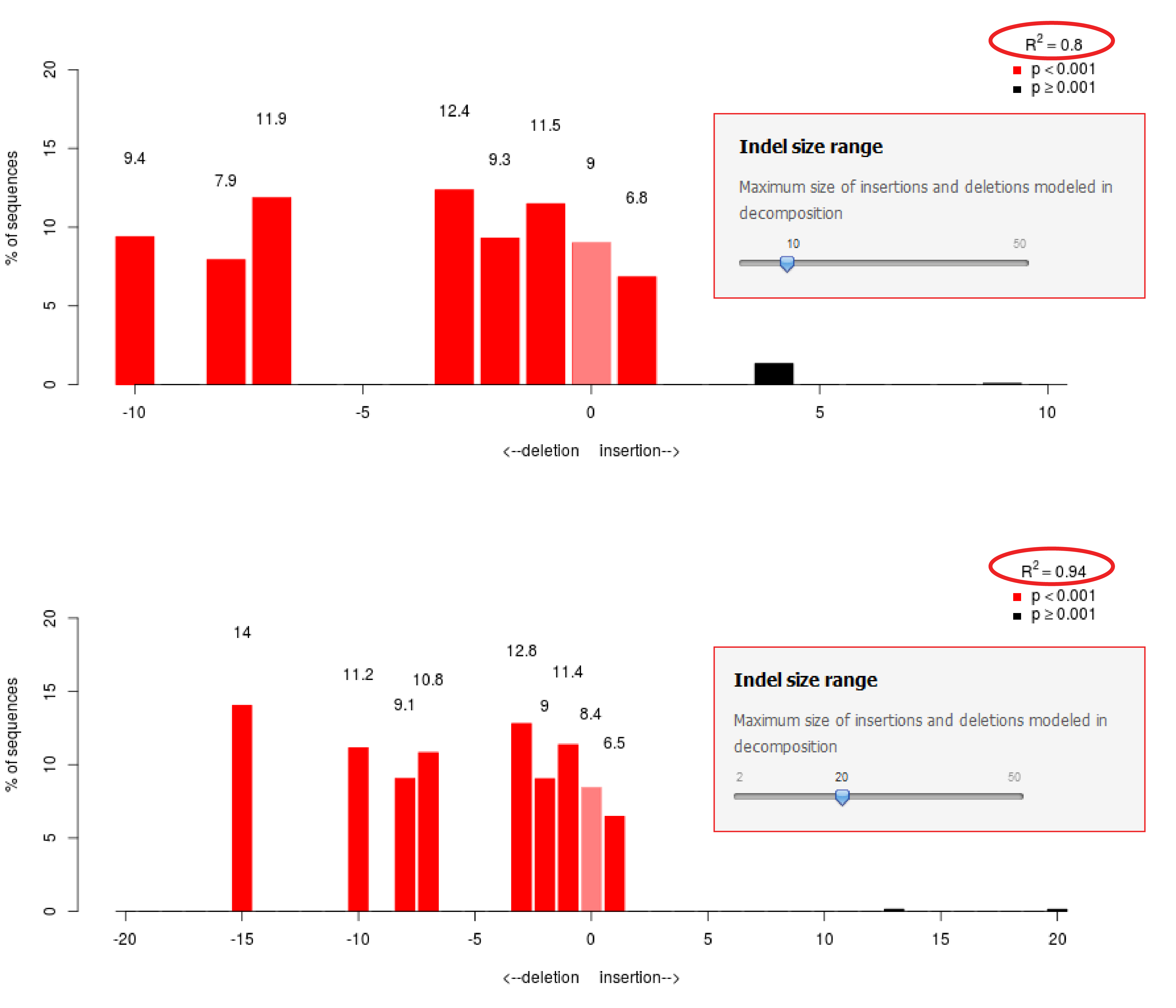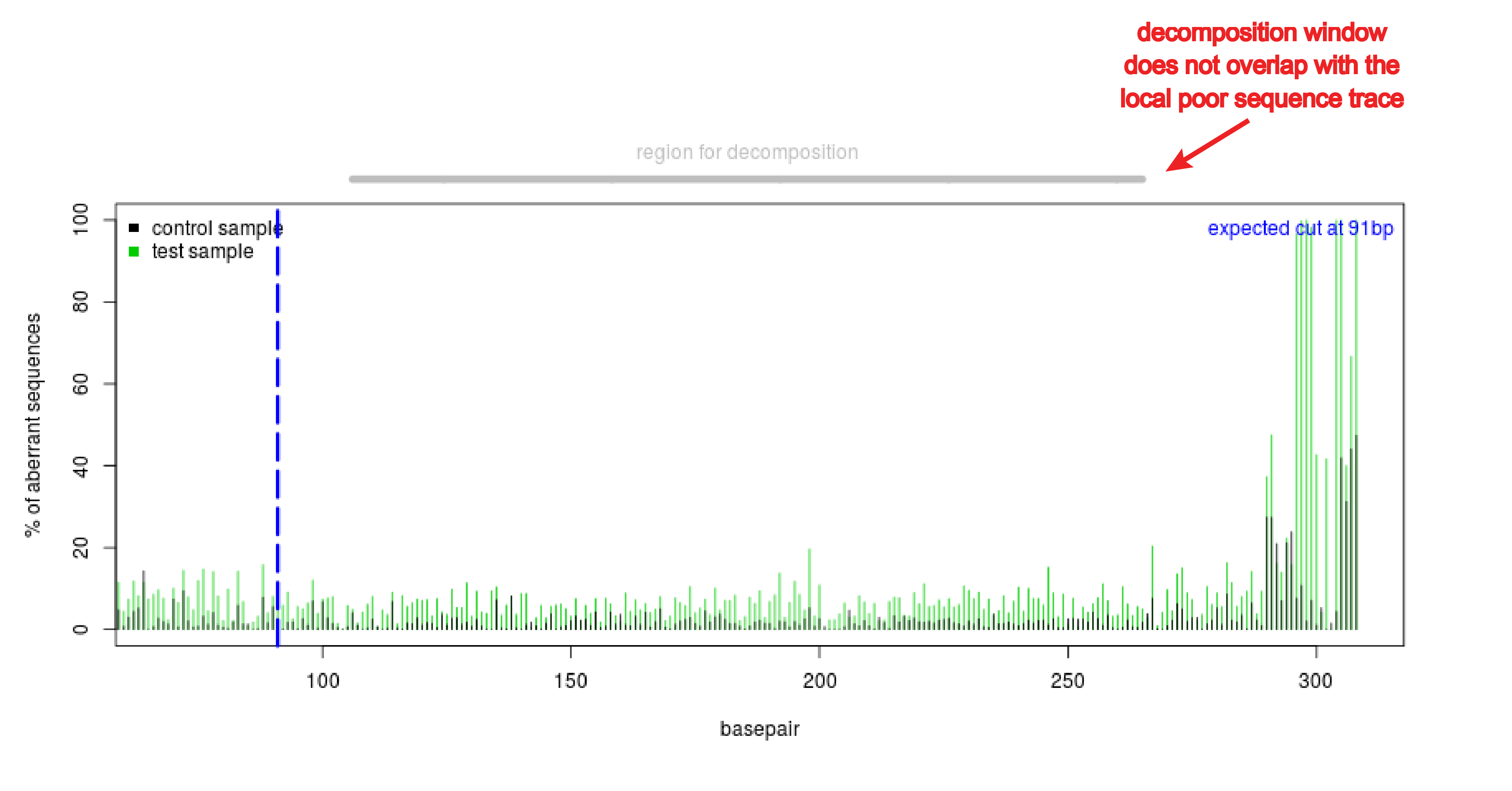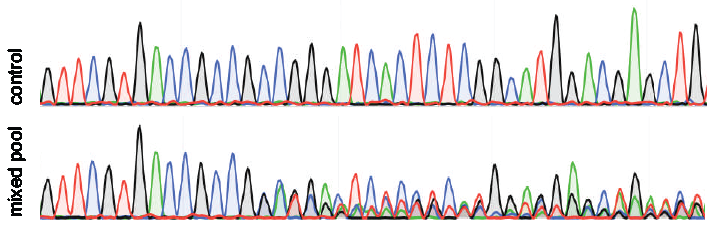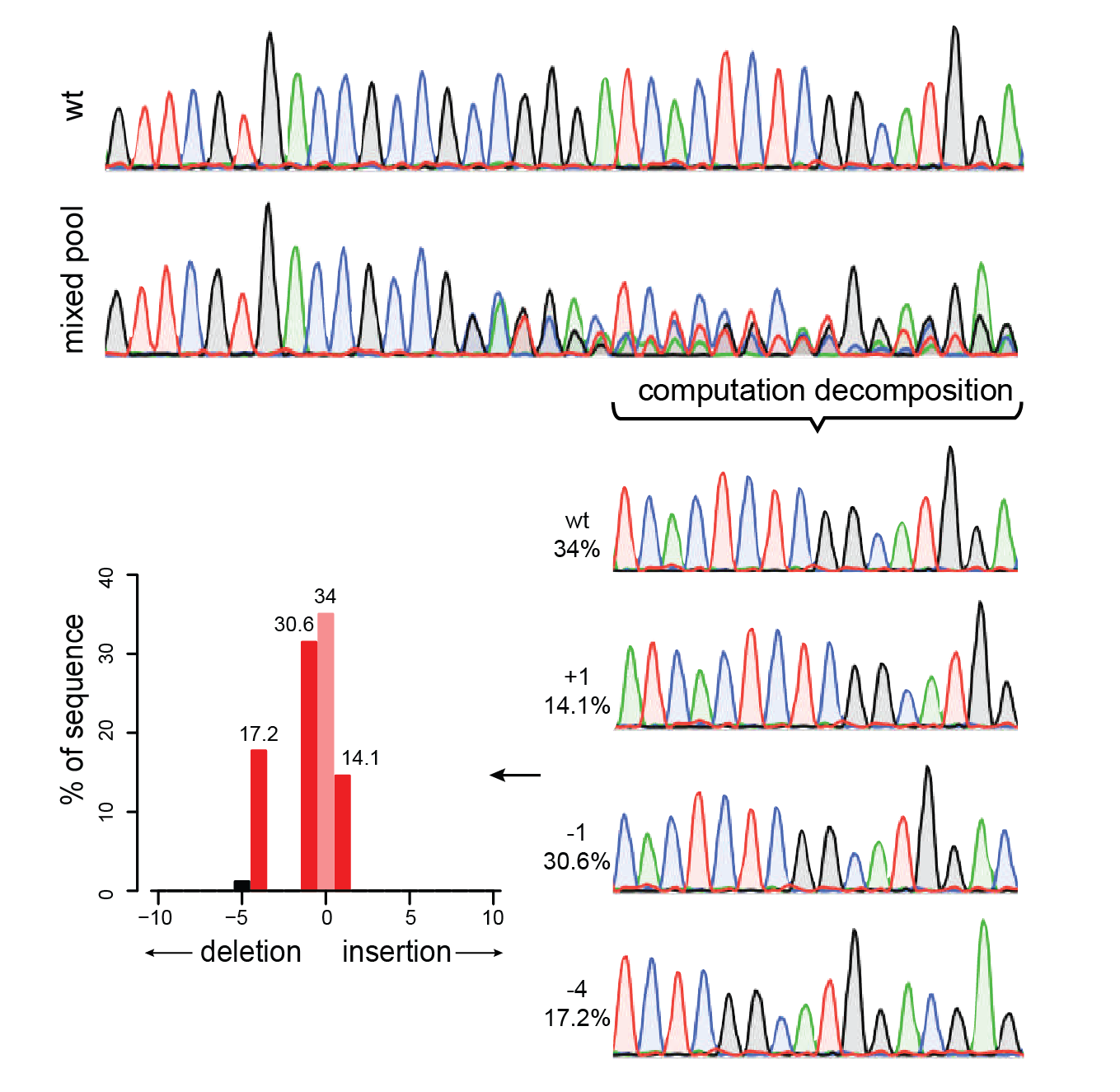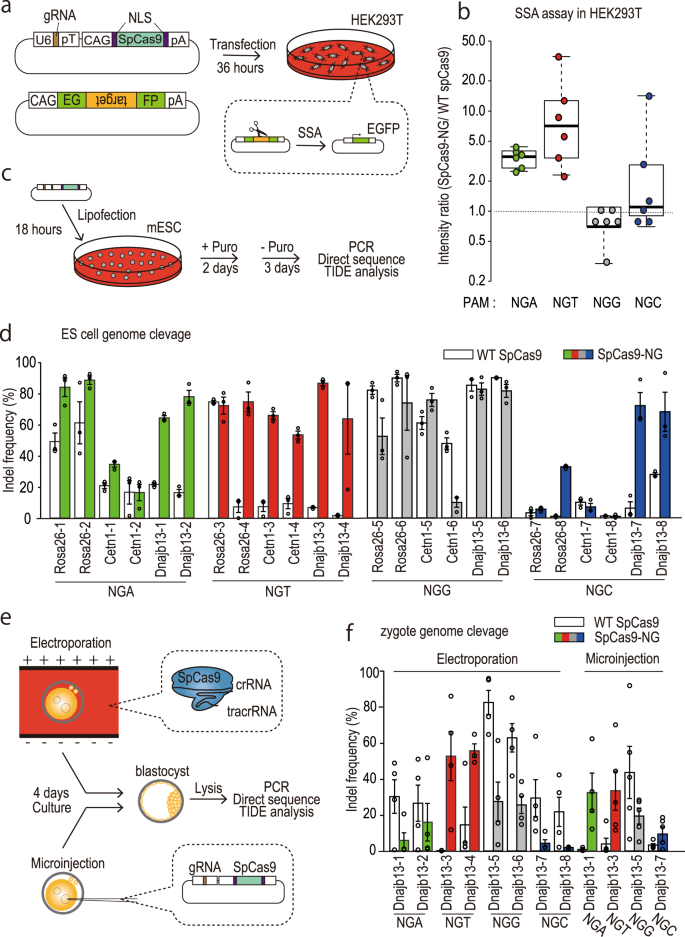
Precise CAG repeat contraction in a Huntington's Disease mouse model is enabled by gene editing with SpCas9-NG | Communications Biology
![Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML](https://www.mdpi.com/cells/cells-08-00574/article_deploy/html/images/cells-08-00574-g004.png)
Cells | Free Full-Text | A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA[Ser]Sec Gene | HTML
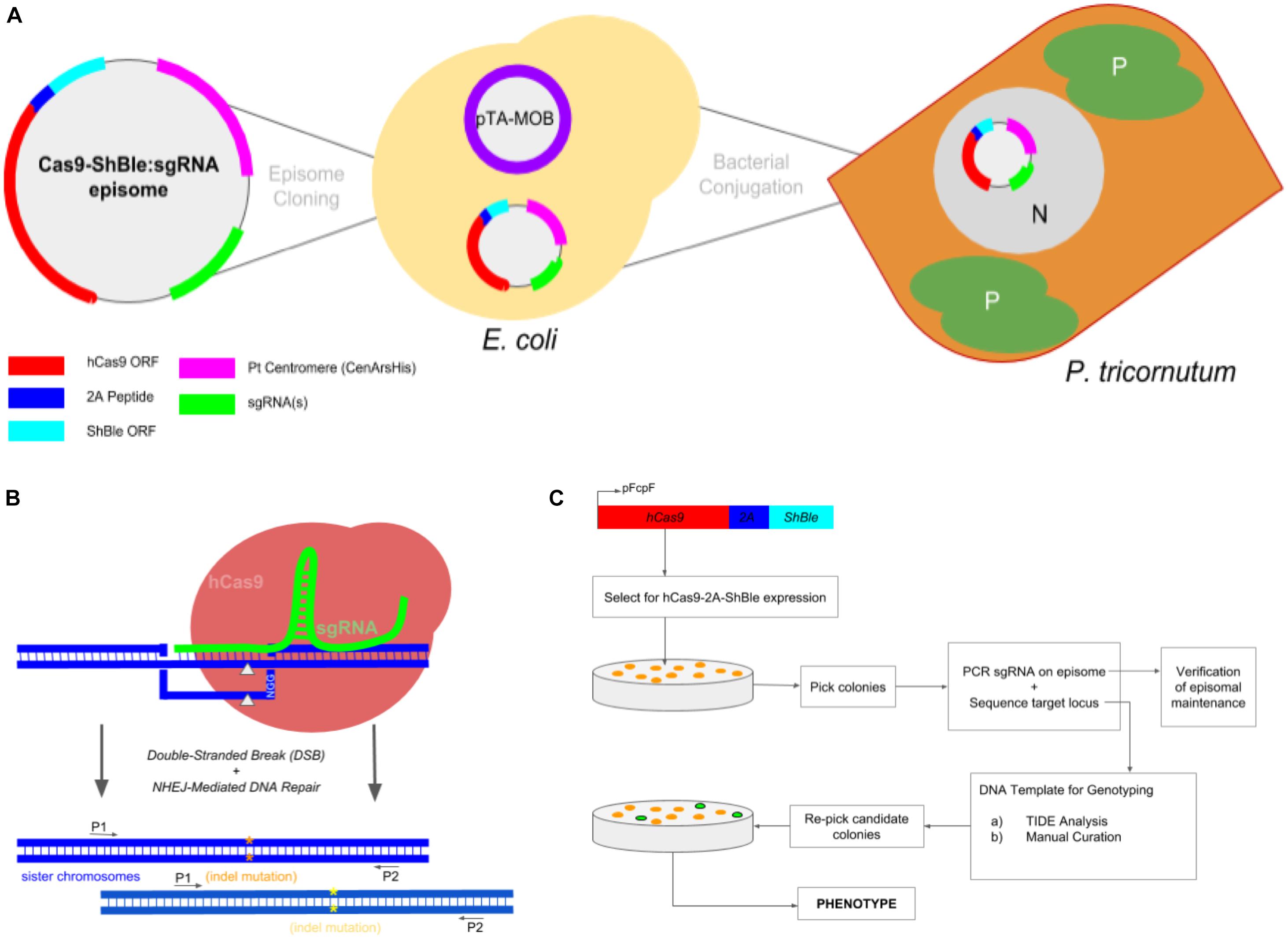
Frontiers | Multiplexed Knockouts in the Model Diatom Phaeodactylum by Episomal Delivery of a Selectable Cas9 | Microbiology
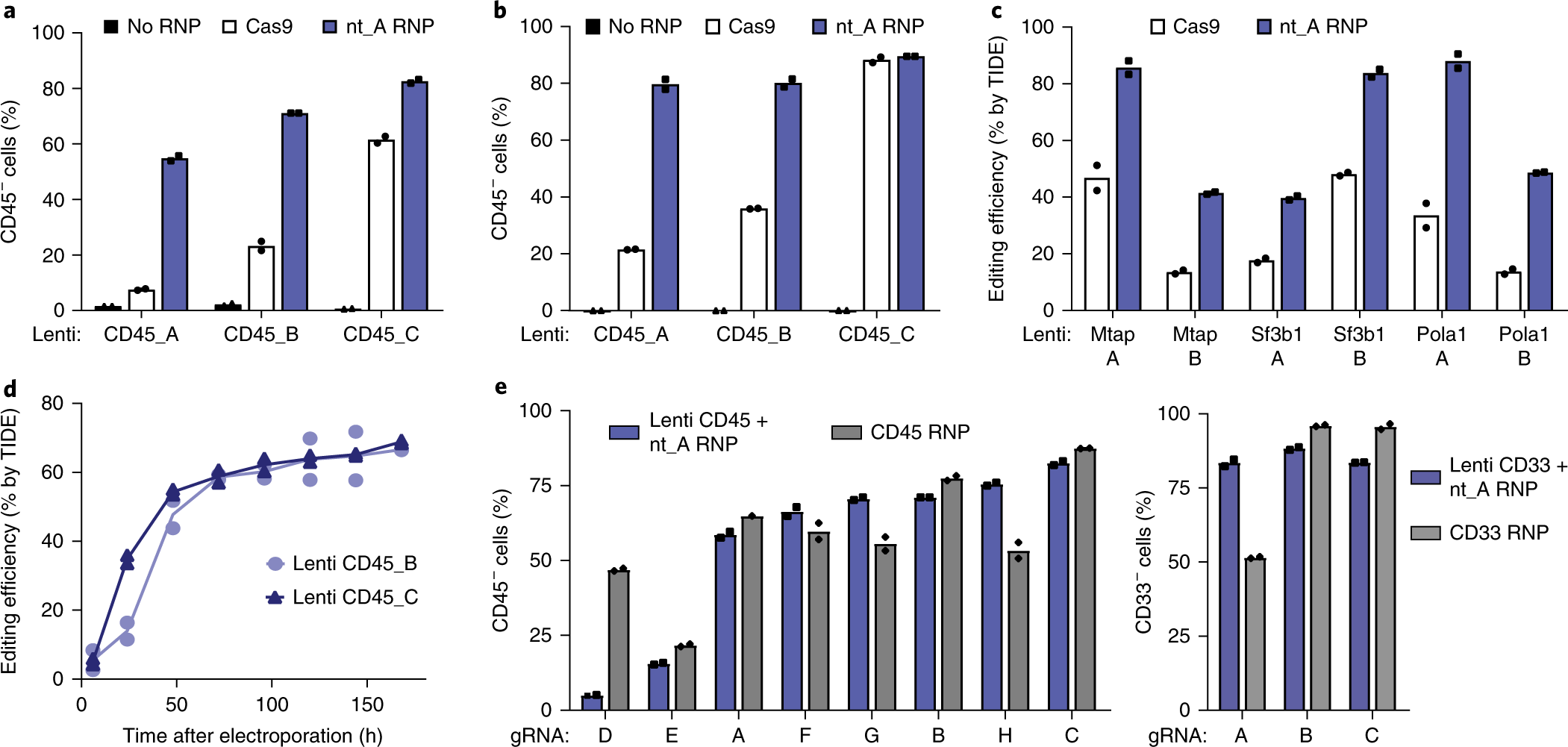
Guide Swap enables genome-scale pooled CRISPR–Cas9 screening in human primary cells | Nature Methods
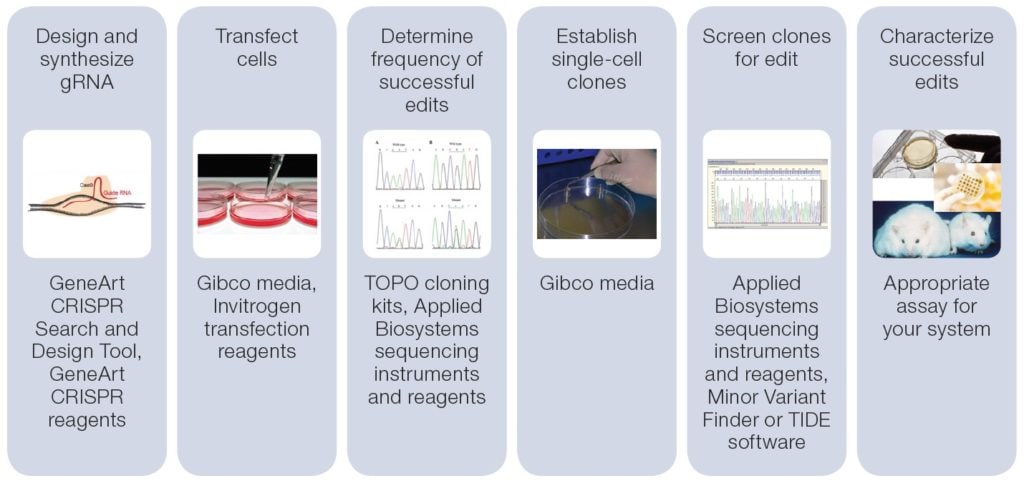
CRISPR-Cas9 Genome Editing Guide – Finessing the technique and breaking new ground - Behind the Bench
An improved strategy for CRISPR/Cas9 gene knockout and subsequent wildtype and mutant gene rescue | PLOS ONE